Abstract:
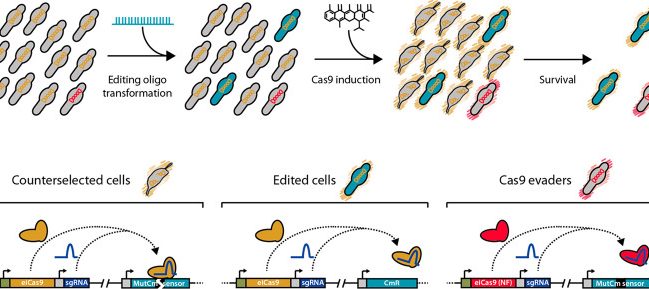
“Mycoplasma species share a set of features, such as lack of a cell wall, streamlined genomes, simplified metabolism, and the use of a deviant genetic code, that make them attractive approximations of what a chassis strain should ideally be. Among them, Mycoplasma pneumoniae arises as a candidate for synthetic biology projects, as it is one of the most deeply characterized bacteria. However, the historical paucity of tools for editing Mycoplasma genomes has precluded the establishment of M. pneumoniae as a suitable chassis strain. Here, we developed an oligonucleotide recombineering method for this strain based on GP35, a ssDNA recombinase originally encoded by a Bacillus subtilis-associated phage. GP35-mediated oligo recombineering is able to carry out point mutations in the M. pneumoniae genome with an efficiency as high as 2.7 × 10-2, outperforming oligo recombineering protocols developed for other bacteria. Gene deletions of different sizes showed a decreasing power trend between efficiency and the scale of the attempted edition. However, the editing rates for all modifications increased when CRISPR/Cas9 was used to counterselect nonedited cells. This allowed edited clones carrying chromosomal deletions of up to 1.8 kb to be recovered with little to no screening of survivor cells. We envision this technology as a major step toward the use of M. pneumoniae, and possibly other Mycoplasmas, as synthetic biology chassis strains.”
Read the full text article.