Mycoplasma team is funded by different European grants supporting projects oriented to achieve a computational whole-cell model that could be used as tool for rational engineering of this microorganism as bacterial chassis for lung therapy or vaccination.
Research lines
Fibrosis
Cancer
Lung cancer is one of the most frequently diagnosed cancers and the leading cause of cancer-related deaths worldwide. This is a heterogeneous disease involving numerous genetic and epigenetic alterations, characterized by presenting wide-ranging clinical and pathological features. It is broadly classified as non-small cell lung cancer (NSCLC, 85% of total diagnoses) or small cell lung cancer (SCLC, 15% of total diagnoses). Lung cancer pathophysiology cannot be fully understood by enumeration of dysregulated pathways in the tumor cells but instead must encompass the contributions of the tumor microenvironment (TME). Host immune cells are a critical component of the TME, and it is well known that are present within it of nearly all premalignant and neoplastic lung lesions. Thus, host immune responses play a dichotomous role throughout the evolution of lung cancer, and can either facilitate the eradication of malignancy or promote an immunosuppressive TME favoring tumor progression. Most lung cancers escape host immune surveillance through a variety of mechanisms. Tumors can directly suppress effector cells by activating negative regulatory pathways (i.e. PD-1) or restrict the effector response through suppression of functional antigen presentation. Lung cancers can also alter TME composition to establish an immunosuppressive milieu characterized by abundant inhibitory molecules, such as TGF-b, and immunosuppressive cells accumulation, including regulatory T cell and myeloid-derived suppressor cells. Current first-line treatments and therapeutic protocols in clinical trials, which have relative efficacy in most cases, are associated with many adverse effects since these are in general systemic therapies. Synthetic biology, which offers innovative approaches for engineering new biological systems, is driving significant advances in biomedicine. Compared to drugs, nanoparticles, or viruses, bacteria provide several advantages when used as a vehicle to deliver therapies. Among others, they contain all biological machinery to synthesize therapeutics and complex regulatory circuits can be integrated to sense and respond specifically to injured tissue. For many years, we have deeply characterized the genome-reduced bacterium Mycoplasma pneumoniae through systems biology analysis. Therefore, it is possible to take advantage of its natural tropism toward the human respiratory tract and solid lung tumours, to integrate genes encoding immunomodulatory molecules within this bacterial chassis. As a result, we will develop strains capable to deliver these specific immuno-active molecules locally, with high bioavailability and fewer systemic adverse effects.
Sensing
In the western world, cancer is the second most common cause of death. The timely detection of micrometastasis is critical for effective treatment and is currently an unfulfilled clinical requirement in the case of lung cancer. To address this issue, our research aims to develop radically new biosensors that draw upon decades of microbial genetics, which have yielded versatile components for engineering bacterial gene expression.
Specifically, we are working to develop and optimize Mycoplasma pneumoniae as a switch-like sensor that can detect metabolites produced in tumour environment for a payload release. These sensors could potentially have a wide range of applications beyond tumour detection in the lung. Additionally, we are designing a sensor that can detect selected cancer-exposed specific antigens.
Screening
Current Grants
Lung-Biorepair
Lung diseases are a leading cause of mortality worldwide. Dysregulation of immunomodulatory molecules plays a key role in many pulmonary diseases, including lung cancer, idiopathic pulmonary fibrosis (IPF) and infections. In IPF acute or chronic inflammation results in senescence of the alveolar cells with telomere shortening and/or dysregulation of miRNAs. Modulating the immune response directly or its downstream repercussions could be a possible way to help treat lung diseases. Systemic treatment with immunomodulatory molecules however, can have several drawbacks and include toxic side effects in other organs, the need for continuous delivery and a high cost of production. Similarly, treating immunomodulatory repercussions such as telomere shortening or abnormal miRNA expression in target cells is not easy due to the lack of a technology that efficiently and specifically delivers RNA. Furthermore, viral transformation can result in toxicity and is associated with high costs. To circumvent these problems, we aim to engineer the genome-reduced lung bacterium Mycoplasma pneumoniae as a vector to locally express immunomodulatory proteins, and/or to deliver protein–RNA complexes into alveolar cells (Mycovector). MPN does not have a cell wall, it directly releases secreted biomolecules into the medium, it does not recombine, it has a unique genetic code that prevents the transfer of genes to other bacteria and we have a non-pathogenic engineered version of it. To design this Mycovector, we will combine our experience in this organism with our know-how in protein design (http://foldxsuite.crg.eu/). We will use our Mycovector expressing different combinations of active biomolecules to treat bleomycin-induced IPF in mice. This project will not only offer new insights into the treatment of a currently incurable disease, but also show that bacterial chassis can be used in other organs different from the gut paving the way to other applications in human health.
Funded by:
European Research Council, Advanced Grant
Novel Engineered cytokines for human therapy – Cytodesign
Cytokines are biomolecules of great potential interest for human therapy. They modulate the immune system and play an important role in cancer, inflammation, immune response and tissue regeneration. Despite their great potential, there are only a handful of cytokines approved for therapeutic purposes. This is because many of them can have adverse side effects as they usually target different cell types1, which adds to their low serum half-life, high production costs, or lack of physiological efficacy. Different methods have been proposed to improve the pharmacodynamics and pharmacokinetics of selected cytokines (e.g. fusion to the IgG-Fc region2 or use of adjuvant polyethylene glycol3). However, other properties do also need improvement, such as achieving effective local concentration at the target site; promoting the right activity in cytokines with dual functionality and decreasing toxicity by removing binding to unwanted cell types. To help solving the issues above we have developed the concept of “Foldikine”, a protein design strategy that can be applied to all helix-bundle cytokines, many of which have been shown to be therapeutically relevant (e.g. IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, IL-2, IL-4, IL-7, IL-9, IL-15, IL-21, GM-CSF, IL6, IL11, IL12A, IL23A, IL27A, IL31, CLCF1, CNTF, CTF1, LIF, OSM, CSF3, IFN1, IFNω, IFNɛ, IFNк, IFNβ, IFNg.
Funded by:
European Research Council, Proof of Concept
Old Grants
MiniCell
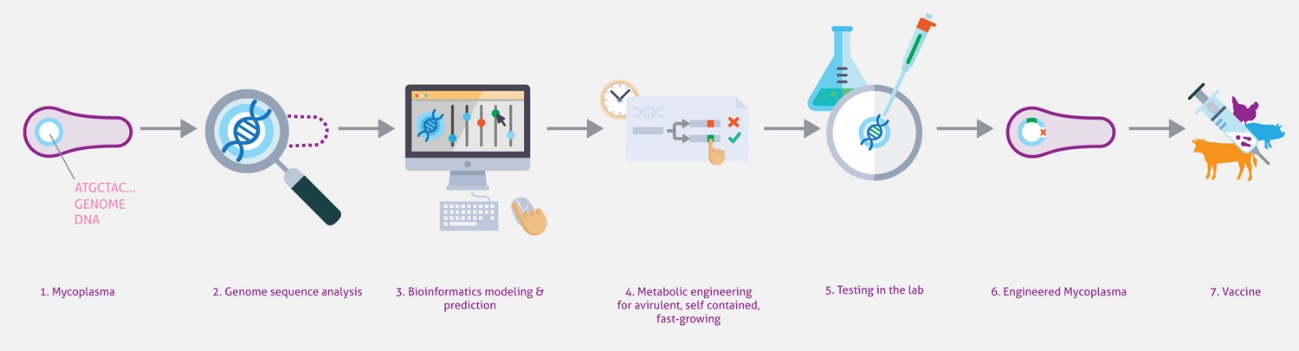
Synthetic biology promises to enable researchers to design therapeutically and industrially valuable organisms. Achieving this promise requires new techniques for designing, synthesizing, and transplanting entire genomes. Here we propose to develop the first model-driven approach to synthetic biology, and use this approach to construct a bacterial chassis capable of synthesizing and delivering human lung therapies in situ. Specifically, we propose to develop a whole-cell model of the human lung pathogen M. pneumoniae, and use this model to design and construct a reduced, non-pathogenic chassis capable of delivering human lung therapies and/or vaccinations. This project will involve intimate integration of predictive modeling, genomic engineering, and systems and synthetic biology. Model predictions will provide direct input into genomic engineering, and the newly created strains will be characterized to refine the computational model. The project will produce the most accurate computational model of any organism to date, as well as produce the most reduced cell to date. In the future we anticipate this reduced chassis could be extended to synthesize and deliver small molecule and/or protein therapies to diseased lungs in situ.
Funded by:

Ref: PCIN-2015-125
MYCOSYNVAC
Official Website
Reference number: 634942
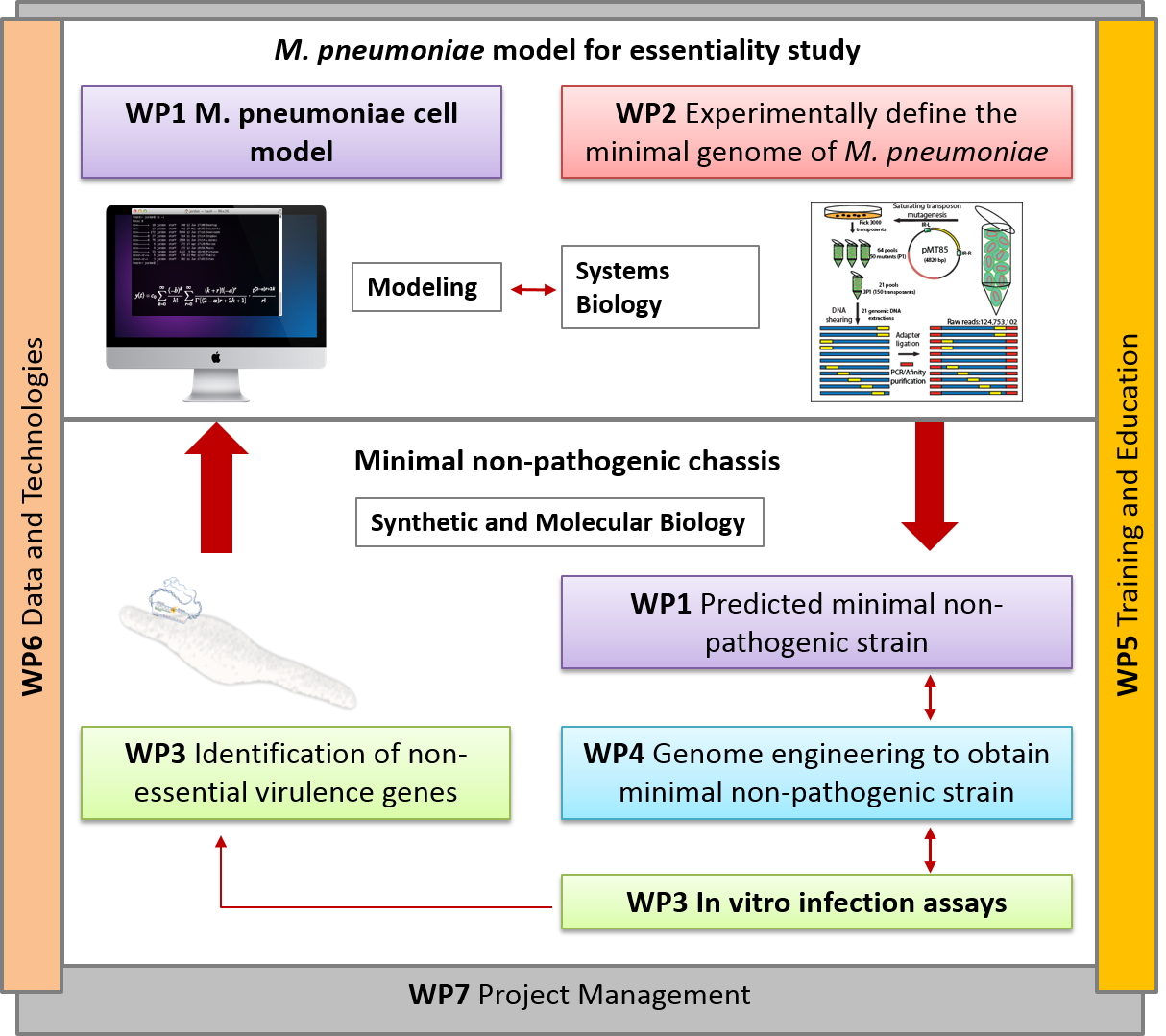
Annually, infections caused by Mycoplasma species in poultry, cows, and pigs result in multimillion Euro losses in USA and Europe. There is no effective vaccination against many Mycoplasmas that infect pets, humans and farm animals (e.g. Mycoplasma bovis cow infection). Furthermore, most Mycoplasmas are difficult to grow in axenic culture, requiring a complex media that includes animal serum. Consequently, even in those cases for which effective vaccines are available (namely, M. hyopneumoniae in pigs and M. gallisepticum and M. synoviae in poultry), the production process of the vaccines is very irreproducible and prone to contamination by animal viruses.
Here we will capitalize on our extensive systems biology knowledge of M. pneumoniae and on cutting-edge synthetic biology methodologies to design a universal Mycoplasma chassis that can be deployed as single- or multi-vaccine in a range of animal hosts. We envision an iterative workflow that is (whole-cell) model-driven and relies on a range of genome-editing and transplantation tools, circuit (re-)design and chassis plug-in as well as on assessment of vaccine performance pigs in an industrial setting. The chassis will be free of virulence determinants from M. pneumoniae and will be optimized for fast growth in a serum-free medium. Using this chassis, we will express heterologous antigens from one or more pathogens (i.e. Mycoplasma and virus) and biological adjuvants to create a targeted vector vaccine. Specifically in this project, we will target the development of attenuated and/or inactivated vaccine(s) against two Mycoplasma pathogens: M. hyopneumoniae (pigs) and M. bovis (cattle), and a combined one against M. hyopneumoniae and PRSSV virus (pigs). We will ensure that foreseeable risks are avoided, all ethical issues are handled in a transparent manner, and that our results and their implications are disseminated effectively and communicated efficiently with the European public.
Funded by:

Ref: 634942
MycoChassis
Engineering bacteria to deliver therapeutic agents or to present antigens for vaccination is an emerging area of research with great clinical potential. The most challenging issue in this field is selecting the right bacteria to engineer into what is referred to as a “chassis”. While the best chassis depends on the application, there are two common drawbacks to bacteria used currently—their complexity and the lack of quantitative information for many reactions—that limits genome engineering to classical trial-and-error approaches. In this project, we want to engineer the genome-reduced bacterium M. pneumoniae using a data-driven, whole-cell model to engineer a chassis for human and animal therapy. Its small size (816 kb), the lack of a cell wall, and the vast amount of comprehensive quantitative omics datasets makes this bacterium one of the best candidates for chassis design. By combining bioinformatics, omics, and biochemistry approaches with genome engineering tools, systems biology analyses, and computational whole-cell models, MycoChassis aims to: i) develop a whole-cell model based on organism-specific experimental data that will be validated experimentally and that can predict the impact of genome modifications; ii) implement genome engineering tools to delete non-essential pathogenic and virulent elements predicted by the whole-cell model to engineer a therapeutical chassis; iii) use the whole-cell model to design and engineer genes and circuits to improve growth rate in a defined medium; iv) as a proof-of-concept, introduce orthogonal gene circuits to secrete peptides and enzymes capable of dissolving in vitro biofilms made by the lung pathogens Pseudomonas aeruginosa and Staphylococus aureus. This project will validate the usefulness of whole-cell models for synthetic biology by modelling multiple genomic modifications orientated to facilitate engineering of biological systems.
Funded by: